FPbase 
A fluorescent protein database
FPbase is a free and open-source, community-editable database for fluorescent proteins (FPs) and their properties. The primary objective is to aggregate structured and searchable FP data that is of interest to the imaging community and FP developers. Each protein in the database has a dedicated page showing amino acid sequence, accession IDs (e.g. GenBank, UniProt), evolution lineages and mutations, fluorescence attributes, structural data, references that introduced or characterized the protein, and more. Excerpts from primary literature can be entered to store key information about a protein that is otherwise difficult to capture within the current database schema. (For more technical information about the structure of the FPbase database, please refer to the database schema page.)
FPbase additionally includes a variety of tools to:
- explore the lineage and directed evolution of FPs
- view relationships between FP properties
- compare spectra with common filters and light sources
- calculate FRET relationships
- create and share curated protein collections
- build a spectra viewer customized for the specifications on your microscope
FPbase was designed and created in 2018 by Talley Lambert at Harvard Medical School.
A Caution About Overinterpretation and Comparison
Fluorescent proteins are tremendously complex! Even "basic" fluorescent proteins demonstrate complex environmentally-dependent variations. As such, any attempt to classify and categorize something as multifacted as fluorescent proteins will innevitably miss important details. Please use the data on this website as the starting point (and not the ending point) in your search for the right protein for your experiments. If there is an aspect of fluorescent proteins that you think should be represented here, let us know!
When comparing protein attributes on this site, remember that all measurements reflect the conditions under which they were performed. Conditions such as temperature, pH, fusion protein, expression level, etc... can all dramatically affect the performance and characteristics of a fluorescent protein and a protein that looks inferior to another based on attributes catalogued in FPbase may actually be superior when used in a particular biological environment. When searching for a fluorescent protein to be used in a biological experiment, it is always advisable to narrow your choices based on a few critical parameters (e.g. spectra), and then test multiple candidate proteins in your biological system.
How to use this site
Search
There are many ways to find and compare fluorescent proteins on this site. Of course, you can always just type the name of the protein you're looking for into the search box on the home page or the top right corner of this page. To perform more complicated searches, use the advanced query builder, where you can search using a number of fields (e.g. Name, Sequence, EC, QY, Spectra, Oligomerization, Year Published, etc...) and a variety of operators (e.g. contains, greater than, etc...):
Browse
If you just want to browse the database without searching something specific, you may find the Table and Interactive Chart views usful.
Table
The table page shows all proteins in the databse, with each row representing a single protein "state" (that is, a multi-state protein such as a photoconvertible protein will appear in multiple rows of the table; multistate proteins have small a parenthetical state name in the protein name). You can sort this table by any of the paremeters, filter based on oligomerization or switching type, or enter text in the search field ot limit the proteins displayed.
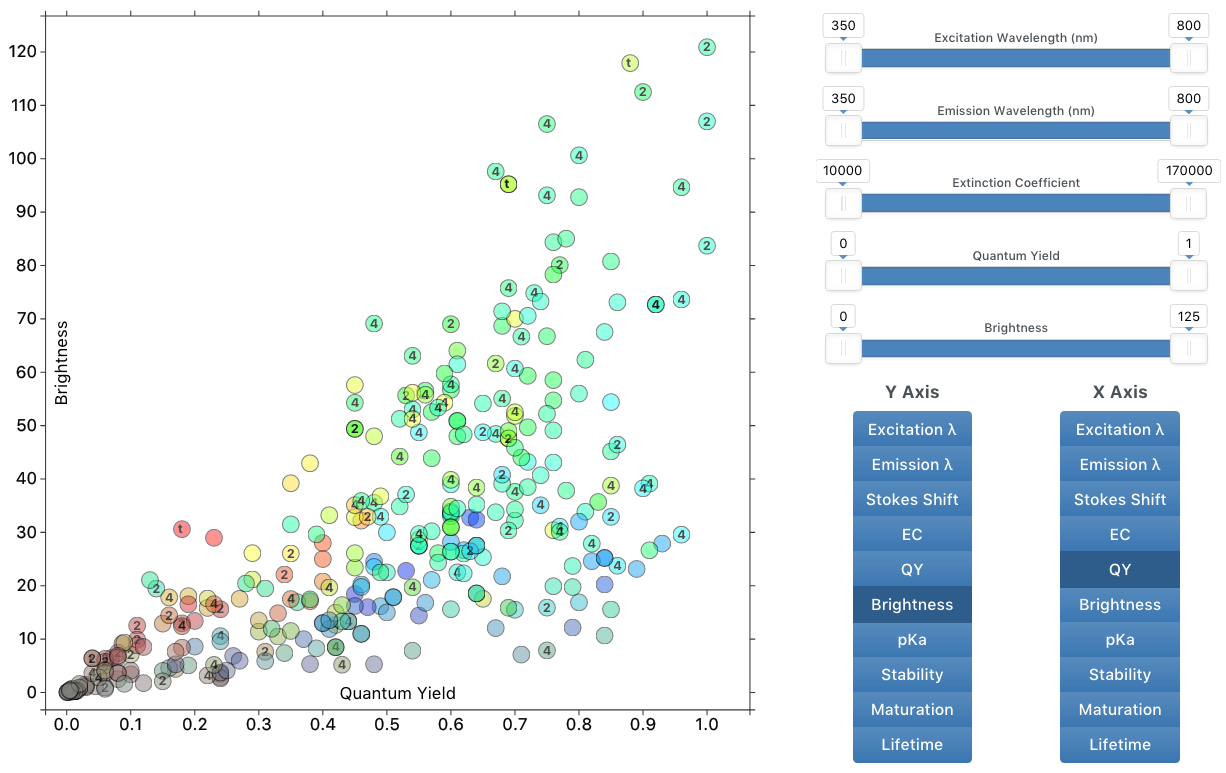
Interactive Chart
The Interactive Chart allows visualization of the many parameters of fluorescent proteins, along with the ability to filter down to a subset of proteins based on certain attributes. Change the axes to explore the relationship between fluorescent protein properties. Hover over the circles to identify each protein on the scatter plot, and click on the circles to go to the FPbase page for that protein.
Interactive Viewer
This page is based on the original GitHub project by Talley Lambert and Kurt Thorn
Collect
Click the heart () icon at the top right of any protein page to quickly add it to your favorites, which can be seen on your profile page. You may also create collections of fluorescent proteins for private use or for sharing publicly. You may view your own collections in your profile, or view all public collections created on FPbase. On each protein page, you can click the book icon () to add the current protein to one of your collections.
Contribute
If you would like to help expand the FPbase database, please consider contributing information to the database. This can be as small as adding an additional reference to a protein, or creating new protein records altogether. Please read the guide to contributing for more information.
REST API
FPbase includes a REST API that allows programmatic retrieval of data in JSON, or CSV format.
Moderation
While we would like for FPbase to eventually be a comprehensive resource for information about fluorescent proteins, we also put a very strong emphasis on the quality and reliability of the data. As such, we aim to moderate all data that is submitted to FPbase. Currently, user-submitted data will be visible on the website immediately, but protein pages that have unmoderated data will be marked as such with the following alert:
Version History
When a change is made to the database, the revision changeset is logged and revertible (by a moderator) for a certain period of time. If the protein has a previously moderated version, you may click on the link to view the previous verision. When viewing an older revision of a protein page, the following alert will be visible at the top of the page:
[date]. Click here to view the current version.
Want to Help?
We have implemented a number of server-side data-integrity checks to help reduce the amount of inaccurate or inconsistent information in the database, but manual curation is still required in many cases. If you believe you would make a good moderator and would like to help the project grow, please contact us. Alternatively, simply adding data to the database is a great way to help the project, and we greatly appreciate it!
